Analysis of protein intact mass
Method overview
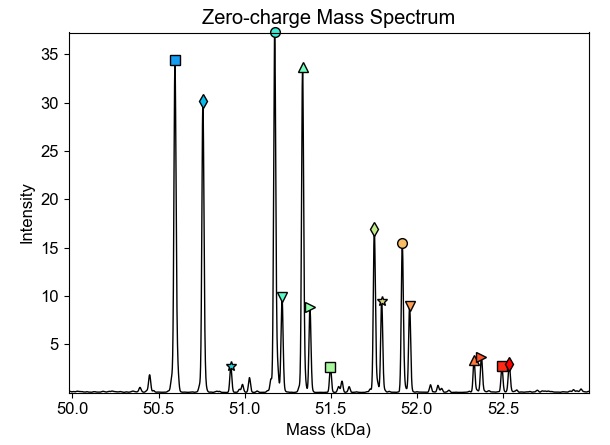
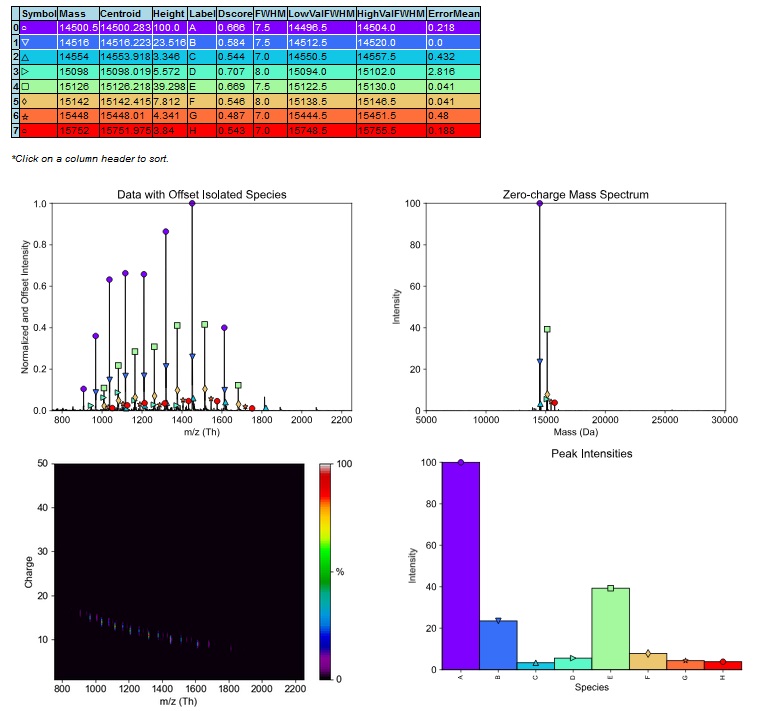
Analysis of intact protein mass using ESI-TOF. This analysis allows for the elucidation and verification of whole protein mass, including analysis of custom covalent post-translational modifications or protein degradation products. It allows for relative quantification of individual proteoforms in the sample. Examples of common applications are antibody-drug conjugate analysis, analysis of methylation of histones or verification of mass and homogeneity of produced proteins, as a more precise alternative to SDS-PAGE.
Contact: František Filandr
Requirements:
- Ideally pure protein in suitable buffer solution
- Any buffer not containing detergents or excessive salt amounts (>250 mM) is usable
- Protein sequence
- List of expected masses of modifications
- Precise buffer composition
Results:
- List of masses found in the sample and their relative ratios. In case of a single pure protein, the result should be a single dominant mass. In case of natural PTM presence, additional signals may appear based on populations of proteoforms in the sample.
- Results in the form of HTML report from UniDec software, showing detected mass, their relative ratios, mass precision and graphical visualization.
- Results in the form of .txt file containing deconvoluted zero-charge mass spectra in coordinate formate. This can be opened for example in mMass software (provided upon request).
- Data interpretation based on information provided by requester in the reQuest system.
Expected turnaround time:
- Within 24 hours
To start an intact protein mass analysis request, use the reQuest system (Link Here)

