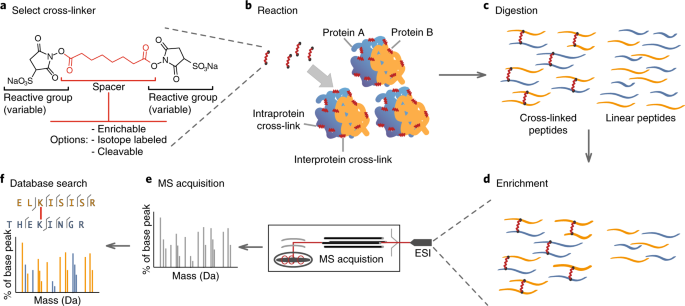
XL-MS (Cross-linking Mass Spectrometry)
This method is based on cross-linking of proteins with bi-functional reagents of specific length targeting specific protein residues and subsequent mass spectrometric detection of formed links. The result is a qualitative information about specific residue distances in individual proteins, or within multiple proteins. This technique can be used for identification and characterization of protein-protein interactions, or residue distances in individual proteins to aid structural modeling.
Contact person: Petra Junková
Requirements:
- Isolated protein or protein complexes in solution
- Sequence of protein(s) with information about existing PTMs
- Selection of cross-linking reagent after discussion with operator
- Suitable buffer for cross-linking reaction. Reagents based on NHS chemistry targeting lysines in proteins (DSBU, DSPU, DSSBU, DSS, BS3, DSSO) react with primary amines, which must not be present in the buffers
- Information about the exact composition of samples, including potential contaminants
Results:
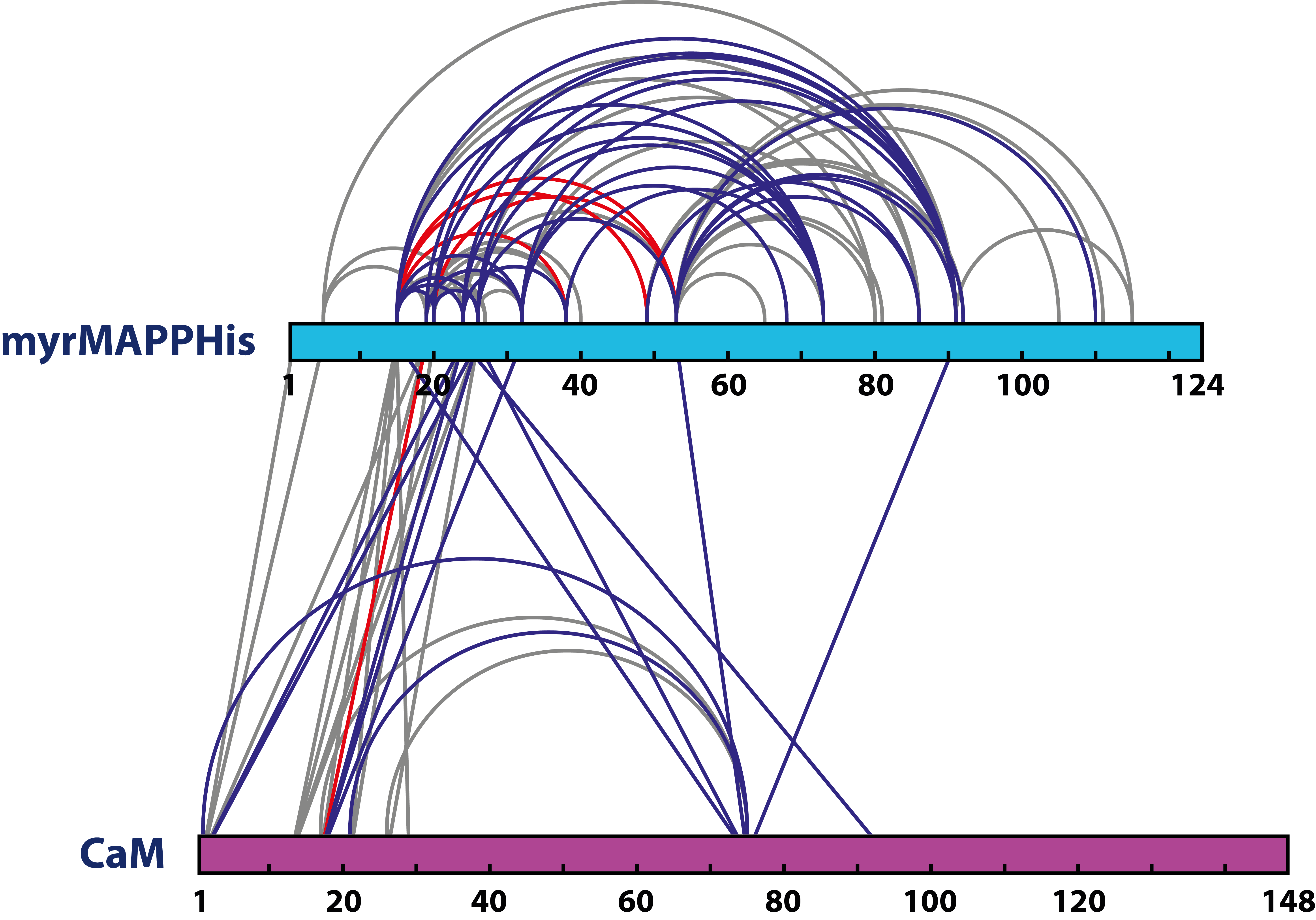
- List of unique cross-links between amino acid residues in analyzes proteins in .xlms or .csv formats
- Schematic illustration of detected cross-links generated from xiView software in vector or bitmap format
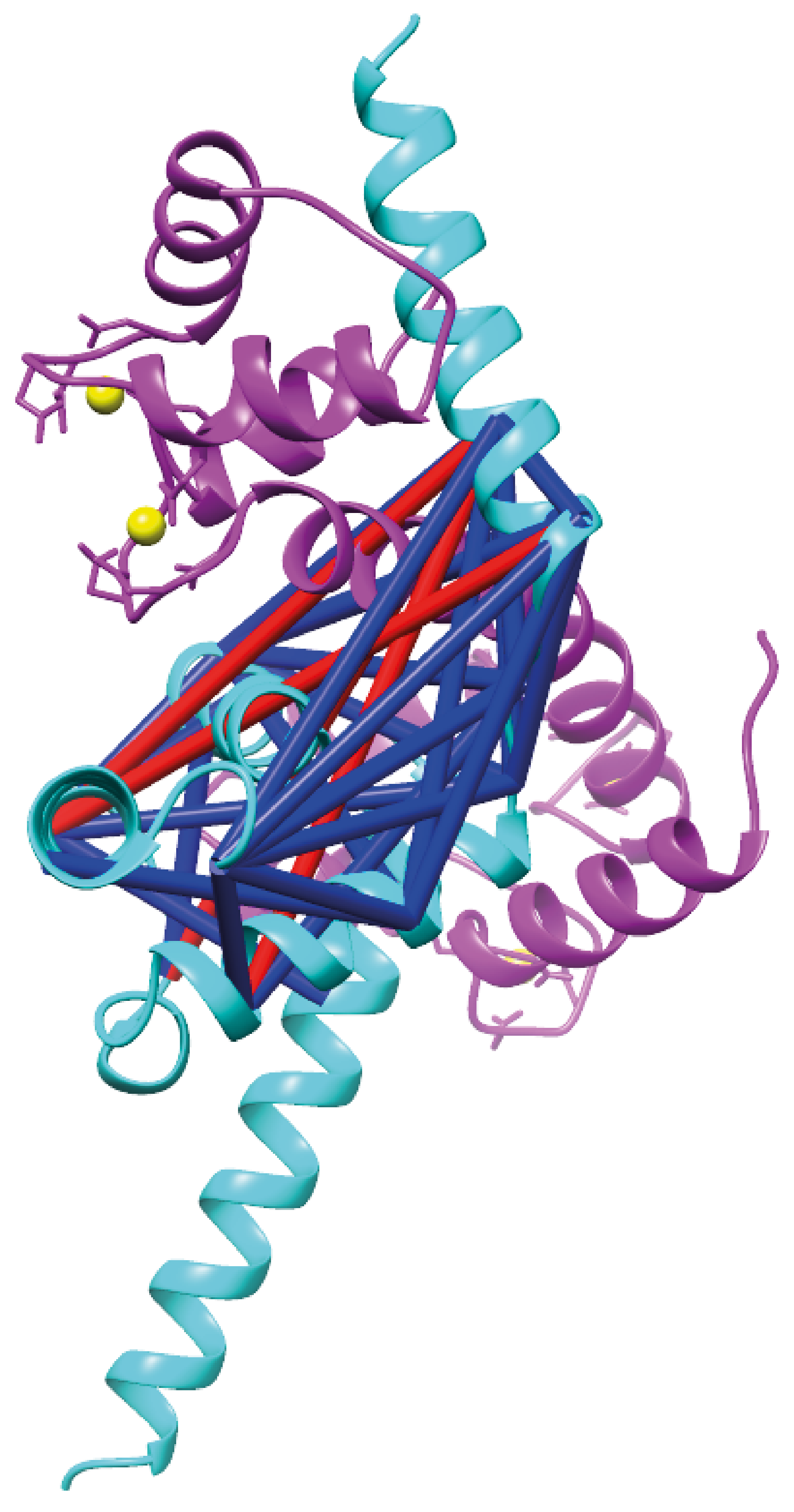
- Validation and visualization of detected cross-links on model protein structures using Xlink Analyzer (UCSF Chimera) in bitmap format, alongside an analysis of structural match between found cross-links and underlying model structure
Expected turnaround time:
- 2 to 3 weeks
If you are interested in XL-MS experiment, please contact us!

